The Tagkopoulos Lab
Software
Please scroll down to access our software tools:
SBROME, Clairvoyance, Mutation Analyser and EVE (Evolution in Variable Environments)
Here you can also access our local databases, source code and datasets
General information and disclaimer
This site contains software packages, code and datasets that was generated by past and current projects. Everything our lab generates is open-source, freely available and we try to publish in open-access venues whenever possible. These tools are provided as a courtecy and are maintained by our students (unfortunately our funding almost never covers software development). If you have any questions, please contact us.
Please select software package or resource from the following.
-
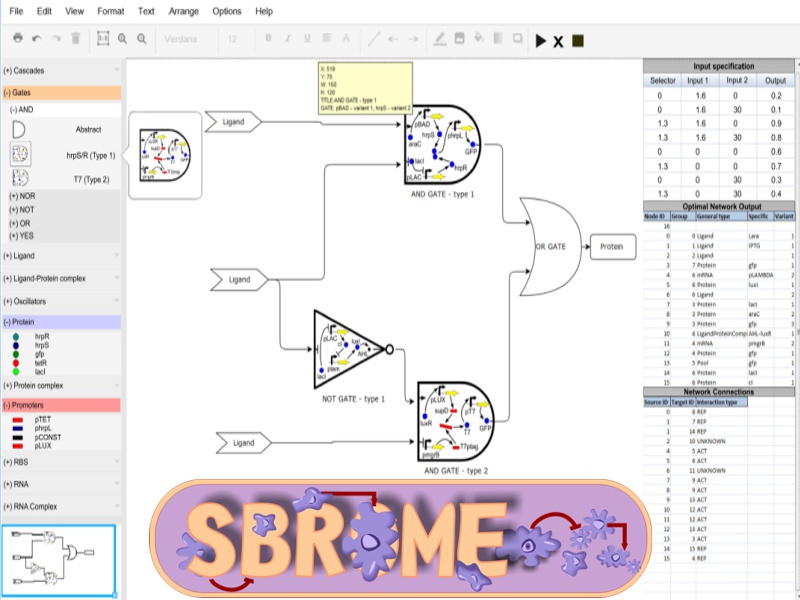
SBROME: A scalable optimization and module matching framework for automated biosystems design
The Synthetic Biology Reusable Optimization Methodology (SBROME) is a CAD optimization platform for modular synthetic circuit design. Given a circuit topology and the desired dynamics, SBROME designs a circuit with the optimal set of parts, minimizing cross-talk and incompatibilities. SBROME is using PAMLib (see resources). -
Mutation Analyzer: A mutation analysis tool
Mutation analyzer loads VCF files containing the mapping of sequencing reads and provides a report on the position and type of mutations. It currently works for E. coli, email us if you want to add another genome. -
EVE: Evolution in Variable Environments
EVE is a multi-scale simulator of microbial evolution. It provides models for genes, proteins, modified proteins, networks, cells, populations. It is written in C++ and it is optimized for HPC runs (scales up to 164K cells) with dynamic loadbalancing, process migration and HDF-5 storage. -
Clairvoyance: A multi-omics bacterial predictor
Clairvoyance is a multi-scale, data-driven system that uses machine learning techniques (artificial neural networks, support vector and constrained regression, optimization solvers) to predict genome-scale gene expression, protein expression and metabolic concentrations, given a specific environmental and genetic background. Specify your strain, genetic perturbations (gene over-expression, knockout), your environment (media, chemicals, stress, other abiotic factors), and Clairvoyance will provide a estimate of growth dynamics and expression levels. Currently only E. coli (Ecomics database) is supported.- Clairvoyance will be accessible after publication
-
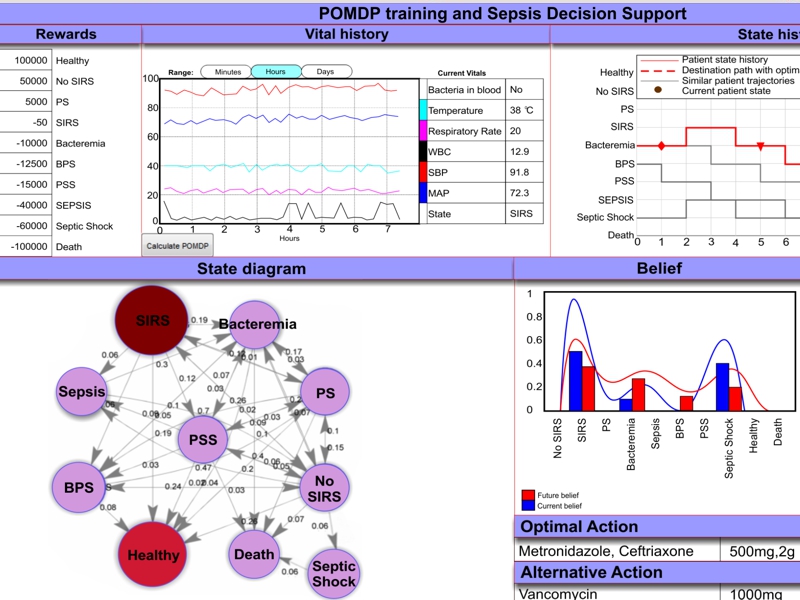
SEPSIS CDSS: A clinical decision support tool.
Sepsis CDSS is a clinical decision support tool that employs probabilistic models trained on Electronic Health Records to inform health decisions. In its latest version, it is based on SVM and POMDP techniques on 1497 patient records from the UC Davis Health System. Sepsis CDSS has also a Google Glass application and supporting user database.