


Patrice Koehl
Department of Computer Science
Genome Center
Room 4319, Genome Center, GBSF
451 East Health Sciences Drive
University of California
Davis, CA 95616
Phone: (530) 754 5121
koehl@cs.ucdavis.edu

 |

| Patrice Koehl |
AIX008: Introduction to Data Science: Summer 2022Application of kNN for classification
The following hands-on exercises were designed to teach you step by step how to build k-NN models based on given data, assess and select the best of thse models based on training data, and finally to use this model to classify test dataset. We use the "Wisconsin breast cancer" dataset (available locally at breast_cancer.csv, which contains information on 569 patients suspected of having breast cancer. For each patient, a biopsy was performed. The actual diagnosis from that biopsy is known (i.e. either the biopsy is "benign" or "malignant". In addition, an analysis of the image of cells from the breast tissue was performed. Ten different features were measured:
For each feature, three values are reported, the mean over all cells in the image, the standard error, and the "worst" value, i.e. the value that differs the most from the mean. The question we would like to answer is: Can we classify the biopsy based on the features of the cells in the image? The breast cancer data setWe first examine the relationships between the features and diagnosis separately. For simplicity, we replace the "B" for benign with 0, and the "M" for malignant with 1. Here is a small Matlab script that reads in the dataset, and illustrates those relationships as scatter plots for three features, Radius, Area, and Fractal dimension. Here is the MATLAB script: read_cancer.m
>> cancer=readtable("breast_cancer.csv");
>> cancer=rmmissing(cancer);
>> figure
>> subplot(1,3,1)
>> subplot(1,3,1)
>> plot(cancer.Radius,cancer.Diagnosis,'or')
>> xlabel('Radius')
>> ylabel('Diagnosis (0:Begnin, 1:Malignant')
>> subplot(1,3,2)
>> plot(cancer.Area,cancer.Diagnosis,'or')
>> xlabel('Area')
>> ylabel('Diagnosis (0:Begnin, 1:Malignant')
>> subplot(1,3,3)
>> plot(cancer.Fractal,cancer.Diagnosis,'or')
>> ylabel('Diagnosis (0:Begnin, 1:Malignant')
>> xlabel('Fractal')
And here are the plots: 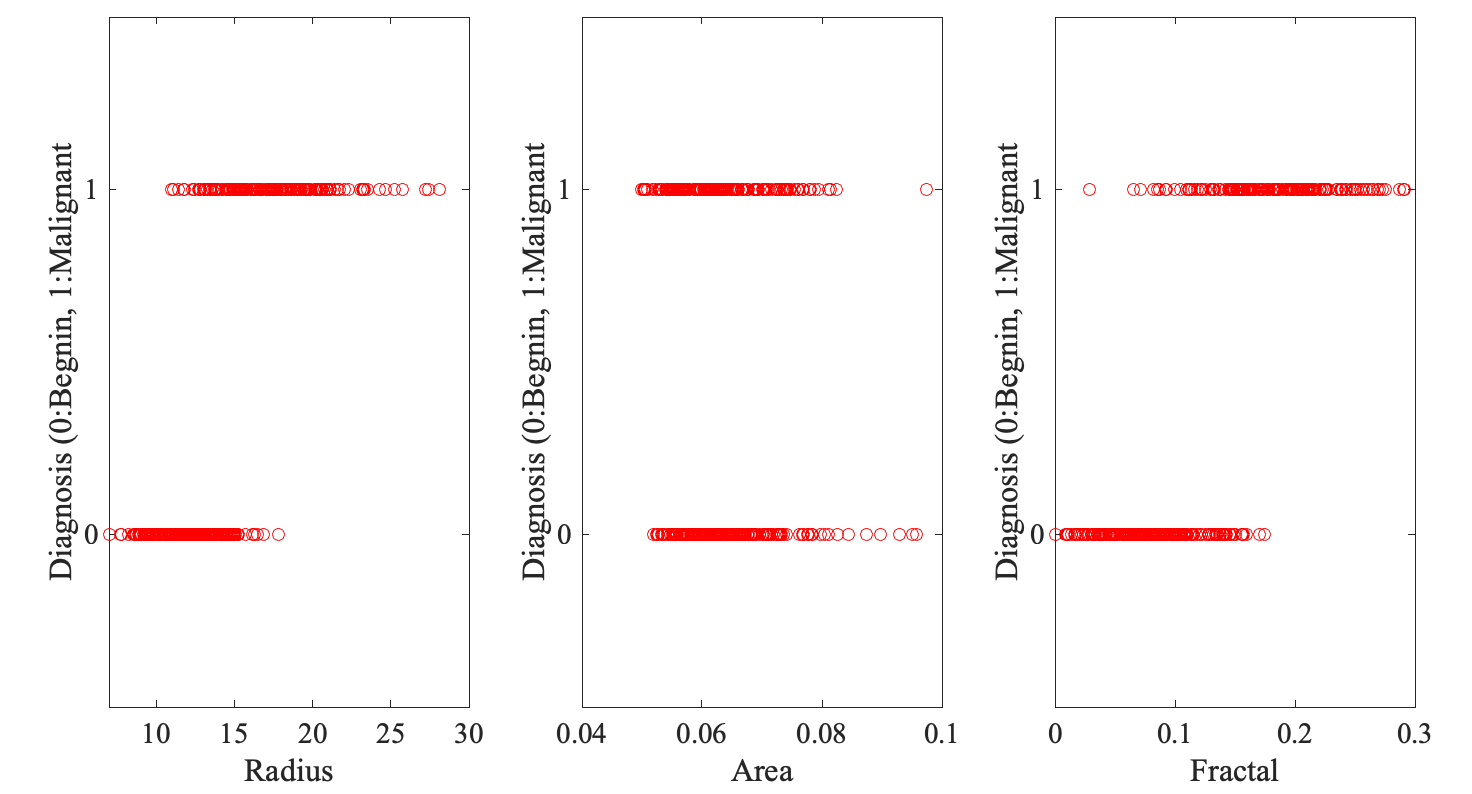
None of the features by itself correlate well with diagnosis, with Radius doing a little better. We will then build first kNN models based on Radius alone. A kNN model to predict cancer diagnosis based on Radii of the cellsOur tast is to build kNN models from Radius alone, choose the best of these models based on a training set, and finally use this model on a test set. To do this, I have divided the breast cancer dataset into three sets:
>> data=readtable("cancer_data.csv");
>> train=readtable("cancer_training.csv");
>> test=readtable("cancer_test.csv");
>> data.Diagnosis=categorical(data.Diagnosis); % Transform diagnosis into categorical variable
>> data.Diagnosis=renamecats(data.Diagnosis,{'B','M'},{'0','1'}); Recast "B" to "0" and "M" to "1"
>> data.Diagnosis=str2double(string(data.Diagnosis)); convert string to number
>> train.Diagnosis=categorical(train.Diagnosis);
>> train.Diagnosis=renamecats(train.Diagnosis,{'B','M'},{'0','1'});
>> train.Diagnosis=str2double(string(train.Diagnosis));
>> test.Diagnosis=categorical(test.Diagnosis);
>> test.Diagnosis=renamecats(test.Diagnosis,{'B','M'},{'0','1'});
>> test.Diagnosis=str2double(string(test.Diagnosis));
>> data_val=[data.Radius data.Diagnosis];
>> train_val=[train.Radius train.Diagnosis];
>> test_val=[test.Radius test.Diagnosis];
>> figure
>> plot(data_val(:,1),data_val(:,2),'ok','LineWidth',0.5);
>> hold on
>> plot(train_val(:,1),train_val(:,2),'ob','LineWidth',1.5);
>> plot(test_val(:,1),test_val(:,2),'or','LineWidth',1.5);
>> xlabel('Radius')
>> ylabel('Diagnosis (0:Begnin, 1:Malignant')
>> ylabel('Diagnosis (0:Benign, 1:Malignant')
>> legend('Data','Train','Test')
And here is the plot: 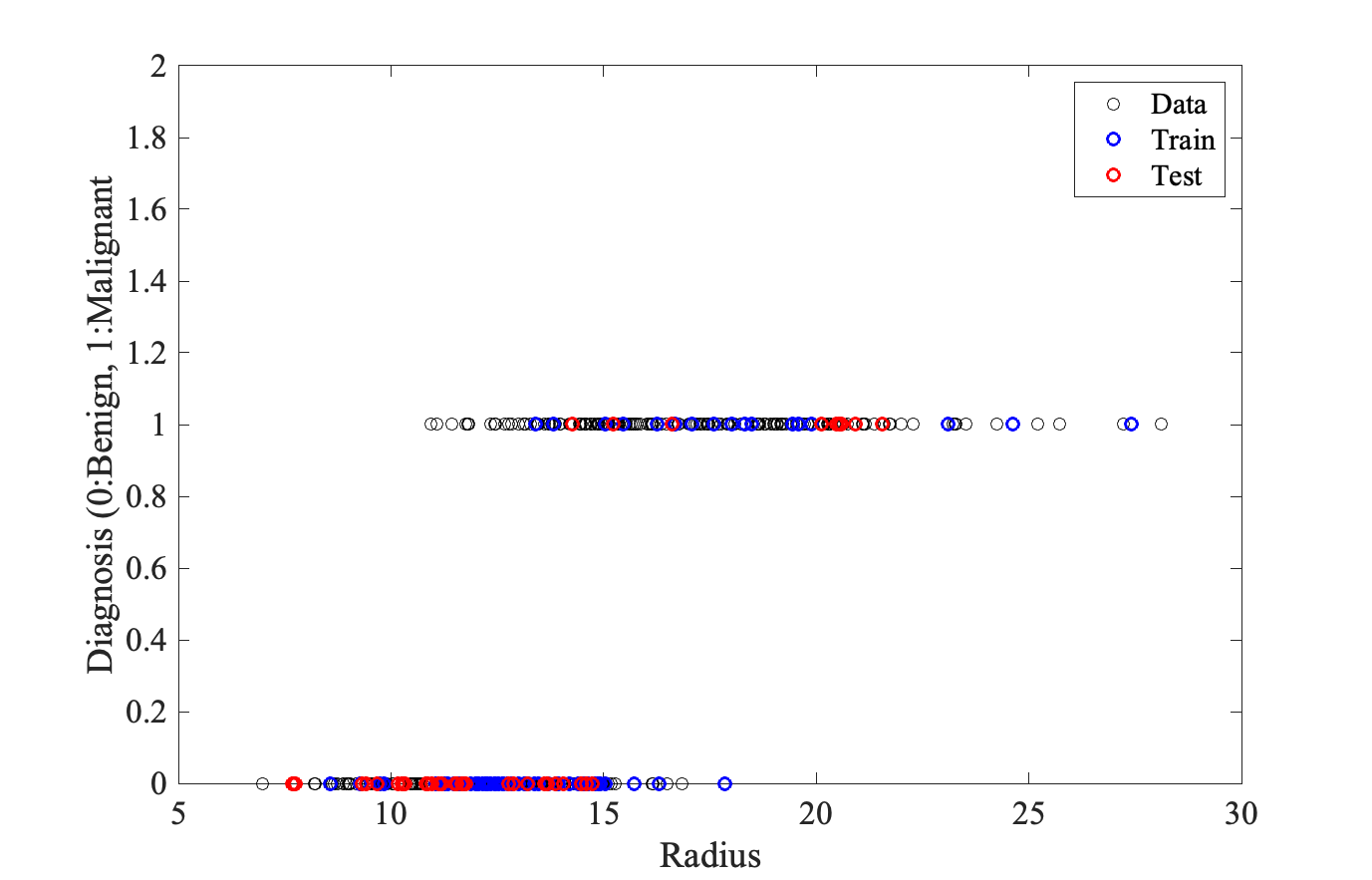
The following Matlab script build a 1-NN model from the Data set and uses the training set to evaluate it using the number of correct prediction (in percentage); It also plots the actual diagnostic and predicted diagnostic for the training set. Here is the MATLAB script: cancer_1NN_radius.m
>> ntrain=max(size(train_val)); % number of training data
>> rmse=0; % Initialize RMSE to 0
>> for i = 1:ntrain % For each training point
val = train_val(i,1); % Radius value for this point
dist=abs(data_val(:,1)-val); % Computes distance to all points in DATA
[t,idx]=sort(dist); % Sort these distances
y=data_val(idx,2); % Order the Diagnosis value in DATA set accordingly
y_predict(i) = y(1); % This is a 1-NN: pick the first value
if train_val(i,2) == y(1) % check if predicted is correct
rmse = rmse + 1;
end
end % end loop
>> rmse = 100*rmse/ntrain; % Compute percent correct
>> figure
>> plot(train_val(:,2),y_predict,'or','LineWidth',1.5);
>> xlabel('Actual diagnostic')
>> ylabel('Predicted diagnostic')
>> title("RMSE = " + rmse);
We find that the 1-NN is "reasonable", with a prediction correct 78.75% of the time on the training set. After adapting this script, you will:
A kNN model to predict diagnostic based on all image featuresThe analysis you have performed above was based on Radius only. Repeat the whole analysis, using now all features. I provide the corresponding script for the 1-NN: cancer_1NN_all.m
>> data_all = [data.Radius data.Texture data.Perimeter data.Area data.Smoothness ...
data.Compactness data.Concavity data.ConcavePoints data.Symmetry data.Fractal data.Diagnosis];
>> train_all = [train.Radius train.Texture train.Perimeter train.Area train.Smoothness ...
train.Compactness train.Concavity train.ConcavePoints train.Symmetry train.Fractal train.Diagnosis];
>> ntrain=max(size(train_all));
>> ndata=max(size(data_all));
>> rmse=0;
>> for i = 1:ntrain
val=train_all(i,1:10);
for j = 1:ndata
dist(j) = norm(data_all(j,1:10)-val);
end
[t,idx]=sort(dist);
y=data_all(idx,11);
y_predict(i) = y(1);
if train_all(i,11) == y(1) % check if predicted is correct
rmse = rmse + 1;
end
end
>> rmse=100*rmse/ntrain;
>> figure
>> plot(train_all(:,11),y_predict,'or','LineWidth',1.5);
>> xlabel('Actual diagnostic')
>> ylabel('Predicted diagnostic')
>> title("RMSE = " + rmse);
This shows that the 1-NN based on ALL data is "reasonable" and better than the 1-NN based on Radius only, with a RMSE of 86.25 %. After adapting this script, you will:
|
| Page last modified 19 September 2024 | http://www.cs.ucdavis.edu/~koehl/ |